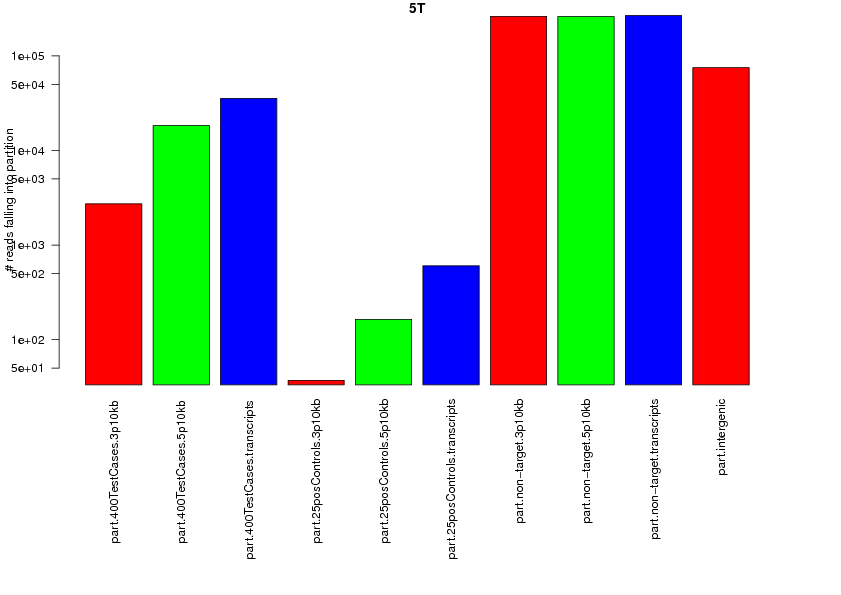
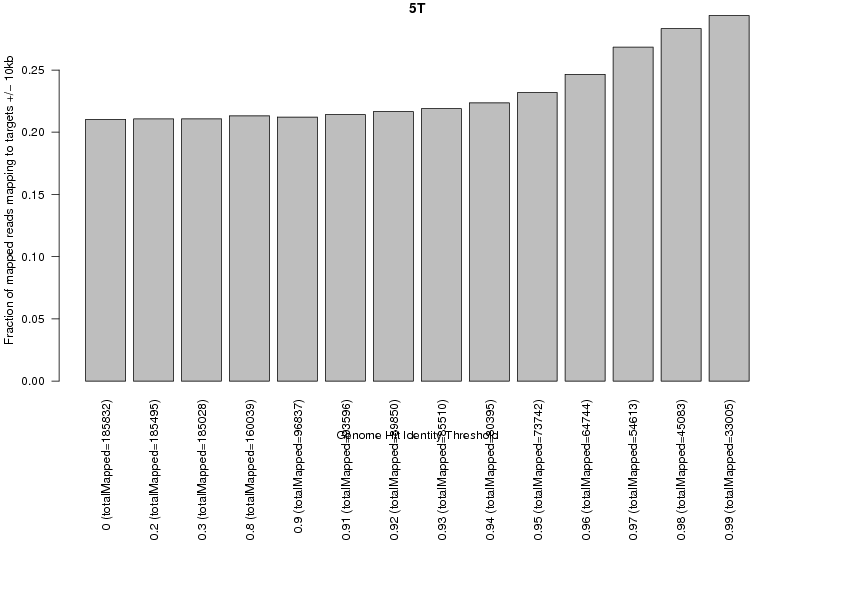
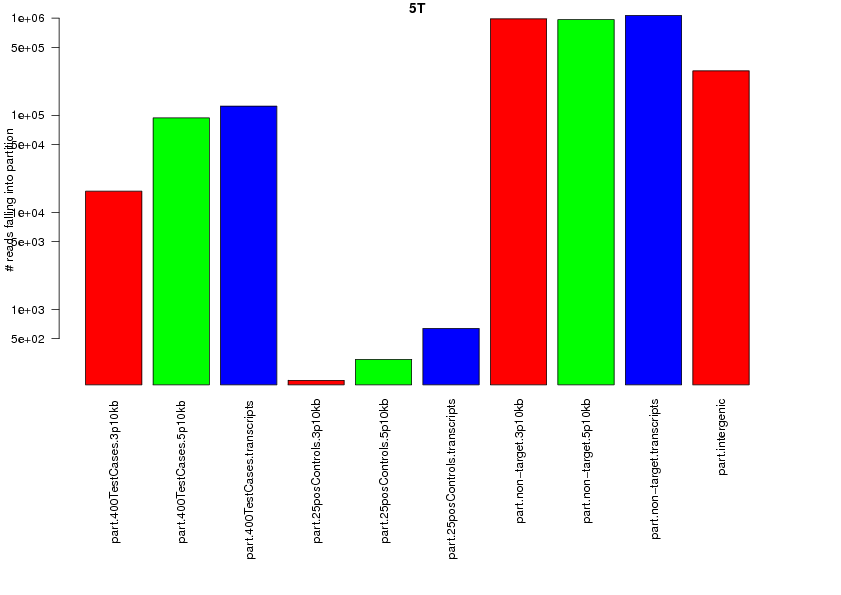
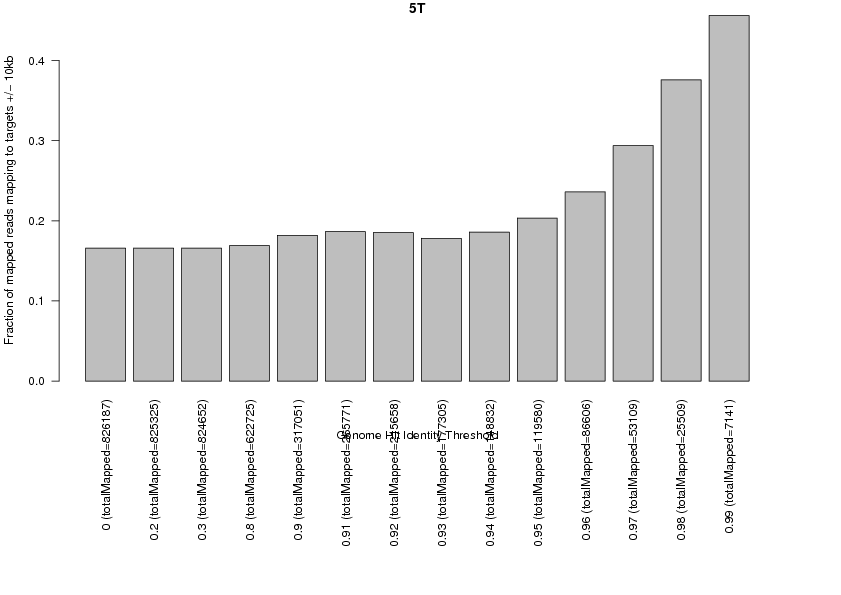
208049 total reads.
189211 (91%) input to Newbler v2.5p1-internal-10Jun23-1.
numAlignedReads = 111377, 58.86%;
numAlignedBases = 33876333, 57.30%;
inferredReadError = 1.57%, 532790;
numberAssembled = 88957; (47%)
numberPartial = 22297;
numberSingleton = 73790; (39%)
numberRepeat = 186;
numberOutlier = 3554;
numberTooShort = 427;
numberOfIsotigs = 4423;
avgContigCnt = 2.0;
largestContigCnt = 16;
numberWithOneContig = 2812;
numberOfBases = 2586937;
avgIsotigSize = 584;
N50IsotigSize = 587;
largestIsotigSize = 3141;
|
845357 total reads.
835080 (99%) input to Newbler v2.6.
numAlignedReads = 589924, 70.64%;
numAlignedBases = 300516134, 69.44%;
inferredReadError = 0.91%, 2744346;
numberAssembled = 504405; (60%)
numberPartial = 85416;
numberSingleton = 214098; (26%)
numberRepeat = 444;
numberOutlier = 22477;
numberTooShort = 8240;
numberOfIsotigs = 17939;
avgContigCnt = 1.9;
largestContigCnt = 14;
numberWithOneContig = 10968;
numberOfBases = 22496014;
avgIsotigSize = 1254;
N50IsotigSize = 1328;
largestIsotigSize = 8287;
|


![[FAIL]](5T_crg_files/tick.png) Basic Statistics
Basic Statistics![[FAIL]](5T_crg_files/error.png) Per base sequence quality
Per base sequence quality
![[FAIL]](5T_crg_files/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](5T_crg_files/error.png) Per base sequence content
Per base sequence content
![[FAIL]](5T_crg_files/error.png) Per base GC content
Per base GC content
![[FAIL]](5T_crg_files/error.png) Per sequence GC content
Per sequence GC content
![[FAIL]](5T_crg_files/tick.png) Per base N content
Per base N content
![[FAIL]](5T_crg_files/warning.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](5T_crg_files/error.png) Sequence Duplication Levels
Sequence Duplication Levels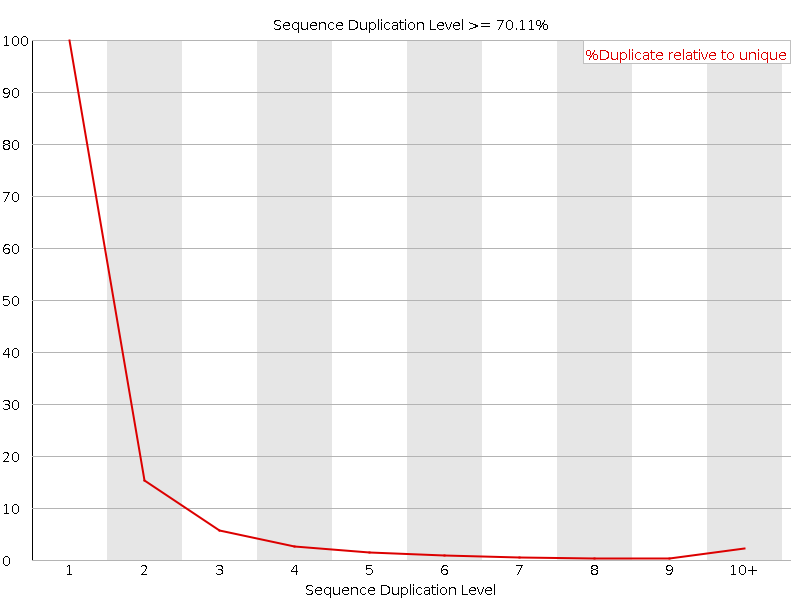
![[FAIL]](5T_crg_files/error.png) Overrepresented sequences
Overrepresented sequences![[FAIL]](5T_Sevilla_files/tick.png) Basic Statistics
Basic Statistics![[FAIL]](5T_Sevilla_files/error.png) Per base sequence quality
Per base sequence quality
![[FAIL]](5T_Sevilla_files/warning.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](5T_Sevilla_files/error.png) Per base sequence content
Per base sequence content
![[FAIL]](5T_Sevilla_files/error.png) Per base GC content
Per base GC content
![[FAIL]](5T_Sevilla_files/warning.png) Per sequence GC content
Per sequence GC content
![[FAIL]](5T_Sevilla_files/tick.png) Per base N content
Per base N content
![[FAIL]](5T_Sevilla_files/warning.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](../../../../../../../../../tmp/5T_fastqc/Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels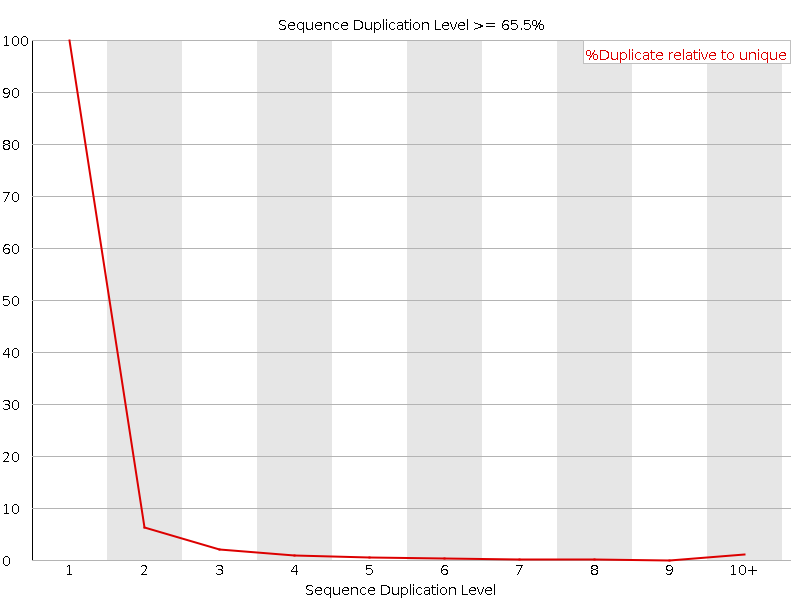
![[FAIL]](5T_Sevilla_files/error.png) Overrepresented sequences
Overrepresented sequences