![[FAIL]](5T_Sevilla_files/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 5T_HEX6GYL01_Amplicons_signalProcessing.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 417922 |
| Sequence length | 40-1596 |
| %GC | 43 |
![[FAIL]](5T_Sevilla_files/error.png) Per base sequence quality
Per base sequence quality

![[FAIL]](5T_Sevilla_files/warning.png) Per sequence quality scores
Per sequence quality scores

![[FAIL]](5T_Sevilla_files/error.png) Per base sequence content
Per base sequence content

![[FAIL]](5T_Sevilla_files/error.png) Per base GC content
Per base GC content

![[FAIL]](5T_Sevilla_files/warning.png) Per sequence GC content
Per sequence GC content

![[FAIL]](5T_Sevilla_files/tick.png) Per base N content
Per base N content

![[FAIL]](5T_Sevilla_files/warning.png) Sequence Length Distribution
Sequence Length Distribution

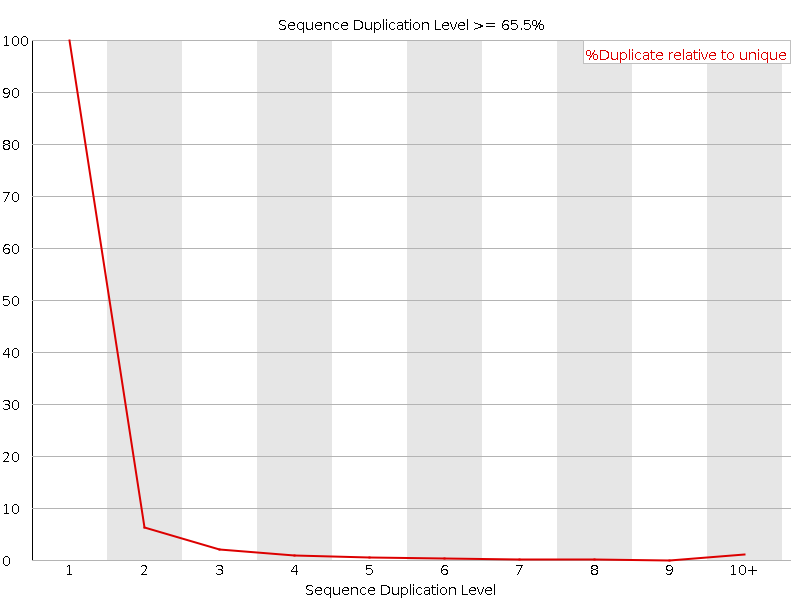
![[FAIL]](5T_Sevilla_files/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](5T_Sevilla_files/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACGCG | 114028 | 27.28451720656008 | No Hit |
| ACCAACGCCAAGAATTAGAGGAGCTGGAAGCAGACTTAAAGAAACGGGAG | 4648 | 1.112169256464125 | No Hit |
| CTAATACGACTCACTATAGGGCTGTGACTAGTATGTTGAGTCCTGTAAGT | 4095 | 0.9798479142040859 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACGTC | 3555 | 0.8506372002431076 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACTTT | 3325 | 0.7956030072597278 | No Hit |
| CGGATTGCAGTCAAGTCCACTGAAGTAGAATAAGGAGACAGGCATCCAGG | 2443 | 0.5845588411234632 | No Hit |
| CTCGGATCAAAACTTTCCCGAGGAAATTTGACAATTTTCTGTGATTTTAT | 1975 | 0.47257622235728197 | No Hit |
| AGTAGGACACCACTTCCAGCAAGAGCAAAAGATGTGTCTTTTTCCTCATG | 1925 | 0.46061226736089506 | No Hit |
| CTAATACGACTCACTATAGGGCTCAGGCGTTTGTGTATGATATGTTTGCG | 1829 | 0.43764147376783225 | No Hit |
| AGTAAGGCTTTAAAGCTCCTGCACGGCGTGCAGTTTTCTGTGTTTGTGTG | 1737 | 0.41562779657448046 | No Hit |
| CGGATTGCAGTCAAGTCCACTGAAGTAGGATAAGGAGACAGGCATCCAGG | 1623 | 0.3883499791827183 | No Hit |
| ATCAACACCTCCATCGGCACGGAATTAGCTCGTTTTCCAGACCGCCATGA | 1543 | 0.3692076511884993 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCACGCAGAGTACGCGG | 1283 | 0.3069950852072875 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGACTTTT | 1155 | 0.27636736041653703 | No Hit |
| CCCTGTTTCTTGCCAACACCTGGTTAGTCTGATTAATTTTAGTCAGTTTA | 1093 | 0.26153205622101733 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATAACGCAGAGTACGCGG | 1071 | 0.25626791602260707 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGTATCAACGCAGAGTACGCGG | 1035 | 0.24765386842520853 | No Hit |
| AAGTGGTGAACTTAGGCATCCTGGCTCCGGGTCATCATCCTCCGAGGAGA | 906 | 0.21678686453453036 | No Hit |
| GAGGGACAAGGTAATTGAGGATGAGGCCATAAACAGCCTCAACTTAGAGA | 830 | 0.1986016529400223 | No Hit |
| TGGTGACTCCAATTAAAGGCCCCAACGTTGTAGGCCCCTACGGGCTACTA | 807 | 0.19309823364168432 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAAGCAGAGTACGCGG | 789 | 0.18879120984298506 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACGTT | 721 | 0.1725202310478989 | No Hit |
| GAAGTCTTCAGGCATGCATGGCTCTCTCTTCATGCTTCCTGGTGGGTCCC | 713 | 0.170605998248477 | No Hit |
| GCGTTCACTGTAGGCCTGAATGCTGATAAAGCTAACCATTTTGGGGTGGT | 658 | 0.15744564775245143 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTACAACGCAGAGTACGCGG | 642 | 0.15361718215360762 | No Hit |
| AATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACGCGGG | 639 | 0.1528993448538244 | No Hit |
| CTAATACGACTCACTATAGGGCAAGGACGCCTCCTAGTTTGTTAGGGACG | 632 | 0.15122439115433023 | No Hit |
| CTAATACGACTCACTATAGGGCAGCAGTGGTATCAACGCAGAGTACGCGG | 588 | 0.14069611075750976 | No Hit |
| CATATCCAGAATTATCCCGCTCCCTGGCGTAGATCGGCTTCTGCTGCCTC | 562 | 0.1344748541593886 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGTTCAACGCAGAGTACGCGG | 549 | 0.131364225860328 | No Hit |
| AGCTGCATTGAGTTAGCGGGGGCAGGCCTCCTAGGGAGAGGAGGGTGGAT | 542 | 0.12968927216083384 | No Hit |
| CGTCACCACCTCCCTCAGCATACAGTTAGCCCGTTTTCCAAACTGCCATG | 538 | 0.12873215576112287 | No Hit |
| GTTCATTGCAAGAGGGTTGGTGTGCCTAGTGTGGGAGTACTTGGTCTGCT | 538 | 0.12873215576112287 | No Hit |
| TAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGTACGCGG | 537 | 0.12849287666119516 | No Hit |
| CAGCAGACCATGTGACTACTGCAGTTGAGAAATCCAAGGAAAGTCAAGTG | 476 | 0.11389685156560315 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCATGGTATCAACGCAGAGTACGCGG | 473 | 0.11317901426581994 | No Hit |
| TATTTGAAGCAGAGGTGGGGCTTCTGTAACGAAAGAGAAAGGAGGAAAAT | 468 | 0.11198261876618125 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGTGGATCAACGCAGAGTACGCGG | 465 | 0.11126478146639804 | No Hit |
| ATTTCCAGAGGAAGGGACCACTTTGGCTTGGTTCGTAAAAAGAGGCAGCA | 457 | 0.10935054866697613 | No Hit |
| AGTTGTGTATTGCTGGGGAGGCCACCGTGAGGCAAATGCTCAGTTTTCGG | 443 | 0.10600064126798779 | No Hit |
| CCAGTGTTAAATGACTGCCACACAGGGTCTGAGTGCTTGCCAGACTTGGC | 441 | 0.10552208306813234 | No Hit |
| CTAATACGACTCACTATAGGGCAAGCAGGGTATCAACGCAGAGTACGCGG | 427 | 0.102172175669144 | No Hit |
![[FAIL]](5T_Sevilla_files/warning.png) Kmer Content
Kmer Content
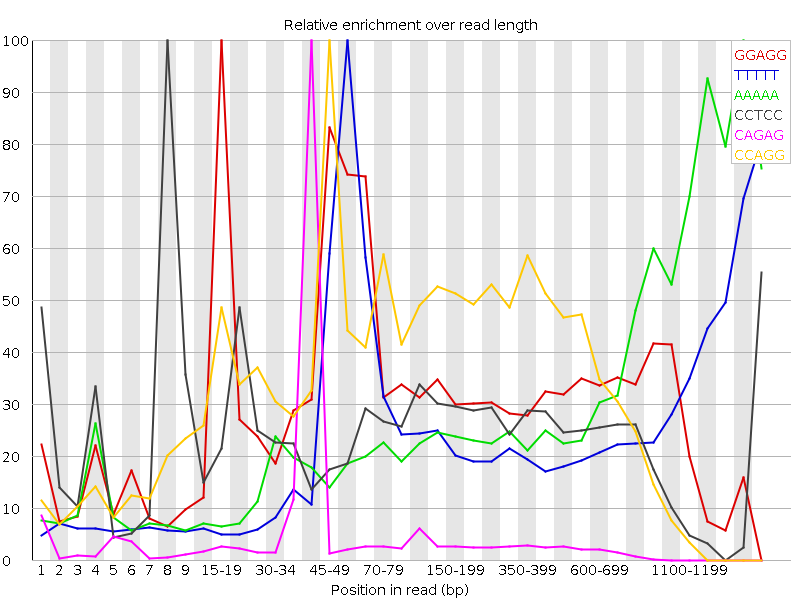
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGAGG | 581240 | 2.5508902 | 7.523727 | 15-19 |
| TTTTT | 1559745 | 2.5310369 | 11.600364 | 50-59 |
| AAAAA | 1509305 | 2.4927402 | 9.323015 | 1400-1499 |
| CCTCC | 507970 | 2.4790454 | 9.352761 | 8 |
| CAGAG | 680220 | 2.4031832 | 80.78036 | 40-44 |
| CCAGG | 504660 | 2.339676 | 5.383005 | 45-49 |
| CCTGG | 504470 | 2.3305647 | 5.1611896 | 90-99 |
| AGCAG | 655655 | 2.3163965 | 89.75112 | 20-24 |
| CTGGG | 512305 | 2.302736 | 5.8099647 | 10-14 |
| CAGTG | 631200 | 2.2221503 | 87.768974 | 25-29 |
| GCTGG | 489235 | 2.1990397 | 8.262544 | 20-24 |
| GGGAG | 479910 | 2.1061828 | 12.116787 | 50-59 |
| GGCTG | 466155 | 2.0952985 | 7.5257335 | 20-24 |
| AGGGC | 461305 | 2.080821 | 141.27472 | 15-19 |
| CAGGC | 441390 | 2.0463471 | 5.13441 | 35-39 |
| GCAGA | 577385 | 2.0398722 | 87.91077 | 35-39 |
| GGGCA | 450760 | 2.0332556 | 122.28088 | 15-19 |
| GCCTG | 438305 | 2.0248938 | 13.5817795 | 1 |
| CTCAC | 540315 | 2.0094354 | 114.160774 | 10-14 |
| AGTGG | 569195 | 1.9496529 | 85.13489 | 25-29 |
| TGCTG | 552290 | 1.9375043 | 5.65888 | 10-14 |
| CTGCC | 404400 | 1.9202037 | 5.4985743 | 1 |
| CTCCA | 516145 | 1.9195471 | 14.921681 | 9 |
| GAGGC | 419465 | 1.8920921 | 5.488826 | 15-19 |
| TGTGT | 706945 | 1.8832618 | 13.393021 | 4 |
| CTGGA | 530125 | 1.8663142 | 5.3230495 | 20-24 |
| GTGGT | 533255 | 1.8201205 | 84.84085 | 25-29 |
| GCAGT | 516080 | 1.8168683 | 85.74486 | 25-29 |
| TGGGG | 407510 | 1.782147 | 7.899591 | 7 |
| TCACT | 618615 | 1.7470171 | 87.92395 | 10-14 |
| TGCAG | 494760 | 1.7418112 | 14.168349 | 6 |
| CCTGC | 362830 | 1.7228178 | 5.014989 | 15-19 |
| AAAAC | 792285 | 1.7171205 | 13.313629 | 1500-1591 |
| TCTCT | 607895 | 1.7107016 | 5.439007 | 6 |
| GGAGA | 497355 | 1.7095969 | 5.816706 | 50-59 |
| CTGAG | 485015 | 1.7075036 | 10.134482 | 3 |
| TAGGG | 495085 | 1.6958053 | 107.206215 | 15-19 |
| AAGCA | 606255 | 1.677582 | 69.18799 | 20-24 |
| CCCTG | 351410 | 1.6685925 | 5.8470755 | 1 |
| GGCCA | 357130 | 1.655706 | 7.1157527 | 20-24 |
| GGGGG | 295255 | 1.6544081 | 30.328264 | 50-59 |
| TCTTT | 770770 | 1.6471004 | 5.1583714 | 8 |
| TTTTA | 1005785 | 1.637876 | 9.828045 | 1500-1591 |
| GGAAA | 608130 | 1.6372482 | 5.281393 | 1500-1591 |
| GGGGT | 371880 | 1.6263279 | 19.603151 | 1300-1399 |
| AGGCC | 349635 | 1.620958 | 7.9025493 | 15-19 |
| CACAC | 430400 | 1.6063136 | 6.2240314 | 2 |
| GGGGA | 363705 | 1.5961934 | 14.2582245 | 50-59 |
| GACTC | 437970 | 1.5847514 | 551.24225 | 8 |
| CCTTT | 556980 | 1.5674195 | 5.060269 | 1500-1591 |
| CACCA | 419685 | 1.5663236 | 7.213267 | 8 |
| GGCAA | 437820 | 1.5467962 | 89.339096 | 20-24 |
| ACACA | 542050 | 1.5416226 | 6.2738132 | 7 |
| GCAAG | 435605 | 1.5389706 | 88.6303 | 20-24 |
| AGGCT | 435605 | 1.5335549 | 12.689636 | 5 |
| TAAAA | 927755 | 1.5268706 | 9.932537 | 1500-1591 |
| GCCAC | 315990 | 1.505707 | 6.0622807 | 20-24 |
| ACTCA | 526770 | 1.4928932 | 431.1039 | 9 |
| TGGTG | 436700 | 1.4905562 | 6.499466 | 1 |
| AGAGT | 552865 | 1.483222 | 64.96143 | 40-44 |
| CAAGC | 407050 | 1.4780719 | 90.43716 | 20-24 |
| CCTGA | 407995 | 1.4762897 | 10.829371 | 2 |
| CTTTC | 519670 | 1.462424 | 6.609426 | 9 |
| CACCT | 392660 | 1.4603053 | 6.4599953 | 6 |
| GTTTT | 701095 | 1.4576788 | 10.642549 | 1500-1591 |
| CCCTT | 388620 | 1.4401947 | 5.229889 | 1400-1499 |
| TTAAA | 877225 | 1.4386294 | 18.480022 | 1500-1591 |
| TTTGG | 537715 | 1.4324425 | 5.050896 | 5 |
| GGGCT | 317700 | 1.4280151 | 15.52505 | 15-19 |
| GGGAA | 414805 | 1.4258415 | 5.3790417 | 50-59 |
| GGCTC | 303315 | 1.4012634 | 6.005629 | 20-24 |
| AAACC | 492630 | 1.401069 | 27.956913 | 1500-1591 |
| ACCCC | 280675 | 1.3746152 | 22.023283 | 1500-1591 |
| GGGCC | 232150 | 1.3741516 | 5.3682756 | 15-19 |
| GCTCC | 289155 | 1.3729883 | 7.2610846 | 15-19 |
| GTACG | 388175 | 1.3665769 | 80.955605 | 40-44 |
| TTGGG | 399525 | 1.3636694 | 6.207198 | 6 |
| ACGAC | 374185 | 1.3587332 | 553.36664 | 6 |
| ACTCC | 362520 | 1.3482146 | 8.977271 | 8 |
| TTTAA | 822850 | 1.3447068 | 19.39451 | 1500-1591 |
| ACCAC | 359355 | 1.3411636 | 7.289356 | 9 |
| GGGTG | 304620 | 1.3321824 | 5.0347867 | 50-59 |
| GGGGC | 231075 | 1.3307873 | 10.191567 | 9 |
| ACCTC | 355880 | 1.3235204 | 6.278024 | 7 |
| CCAAG | 364185 | 1.3224212 | 16.257198 | 8 |
| CTCTT | 468640 | 1.3188183 | 5.370158 | 7 |
| TGGTA | 489855 | 1.3095547 | 65.70016 | 25-29 |
| CCCAA | 350320 | 1.3074439 | 6.756651 | 4 |
| AAGGG | 380300 | 1.3072346 | 5.880659 | 1100-1199 |
| AGTAC | 472915 | 1.304009 | 66.78338 | 40-44 |
| GTAGG | 375685 | 1.2868267 | 6.578207 | 2 |
| AGTAG | 472880 | 1.2686391 | 5.414869 | 1 |
| CCCCC | 196315 | 1.2616832 | 15.871988 | 1400-1499 |
| AGGGT | 367895 | 1.2601438 | 6.080069 | 1500-1591 |
| ACCCA | 336305 | 1.2551377 | 6.7566495 | 3 |
| GCGGG | 216580 | 1.2473089 | 129.68411 | 45-49 |
| CACTA | 436930 | 1.2382821 | 87.91089 | 10-14 |
| GTCTC | 336885 | 1.2146956 | 6.2042403 | 5 |
| GGTAG | 353705 | 1.211539 | 5.3657036 | 1000-1099 |
| GGGTA | 352955 | 1.2089701 | 17.09109 | 1400-1499 |
| GGTTT | 453060 | 1.2069265 | 17.651785 | 1500-1591 |
| GGGTT | 353080 | 1.2051423 | 25.446424 | 1500-1591 |
| AGCAA | 434005 | 1.2009451 | 7.5821667 | 6 |
| TTTAC | 551550 | 1.1827996 | 7.638496 | 1200-1299 |
| AATAC | 547670 | 1.1827891 | 329.94324 | 3 |
| TCAAC | 417275 | 1.1825788 | 68.55018 | 30-34 |
| AAGGC | 334680 | 1.1824076 | 12.446339 | 4 |
| CAAGA | 426910 | 1.1813123 | 14.250411 | 9 |
| AGGAC | 334065 | 1.1802349 | 6.727361 | 4 |
| CAAGG | 333645 | 1.1787511 | 5.690235 | 7 |
| AACCC | 315595 | 1.1778451 | 25.767855 | 1500-1591 |
| AACCT | 414430 | 1.1745158 | 8.106871 | 1500-1591 |
| GAGTA | 436470 | 1.1709584 | 64.94611 | 40-44 |
| GCACA | 321515 | 1.1674789 | 5.892794 | 2 |
| CAACA | 409750 | 1.1653535 | 8.349623 | 8 |
| ACTAT | 540815 | 1.1638743 | 66.93707 | 10-14 |
| GTAAA | 551495 | 1.1588298 | 8.95426 | 1300-1399 |
| GGGAC | 255615 | 1.1530095 | 5.259744 | 50-59 |
| CGGGG | 200055 | 1.1521394 | 62.283924 | 45-49 |
| CGCAG | 247955 | 1.1495548 | 114.96465 | 35-39 |
| TGAGC | 325385 | 1.1455235 | 9.890451 | 4 |
| CCAAC | 306540 | 1.1440506 | 16.830755 | 2 |
| ATAGG | 425995 | 1.1428564 | 83.34604 | 15-19 |
| GGTGA | 333230 | 1.1414064 | 5.2374253 | 2 |
| ACCTT | 401670 | 1.1343473 | 6.3065505 | 1500-1591 |
| TTGGT | 424690 | 1.1313503 | 5.656706 | 1500-1591 |
| GAGCA | 317855 | 1.1229658 | 9.882163 | 5 |
| TGGTT | 420695 | 1.1207079 | 22.143934 | 1500-1591 |
| GCCAA | 306635 | 1.113447 | 16.33123 | 7 |
| GGCTT | 315665 | 1.1073934 | 12.273216 | 6 |
| GCTTT | 403210 | 1.1039938 | 9.478478 | 7 |
| CTTTA | 511475 | 1.0968589 | 7.1877627 | 8 |
| CTAAT | 508400 | 1.0941148 | 329.46176 | 1 |
| ACCAA | 382315 | 1.0873266 | 13.104266 | 1 |
| AGACC | 297945 | 1.081892 | 6.366663 | 8 |
| TAAAC | 495180 | 1.0694277 | 17.22038 | 1500-1591 |
| GGACA | 302525 | 1.0688057 | 7.447636 | 5 |
| CAGTC | 291870 | 1.0561029 | 13.750809 | 8 |
| ATCAA | 485790 | 1.0491483 | 53.011505 | 30-34 |
| TACCC | 281400 | 1.0465287 | 13.68322 | 1400-1499 |
| GGTAT | 391385 | 1.0463097 | 63.089104 | 30-34 |
| AGTCA | 378795 | 1.0444839 | 11.366957 | 9 |
| TTACC | 367125 | 1.0367895 | 13.056303 | 1300-1399 |
| CTATA | 480445 | 1.0339534 | 66.67732 | 10-14 |
| GACCA | 282660 | 1.0263895 | 6.3962755 | 9 |
| AGTAA | 487595 | 1.0245599 | 7.6425543 | 1 |
| CGACT | 282665 | 1.0227954 | 551.28644 | 7 |
| ACACC | 271715 | 1.0140787 | 7.000217 | 7 |
| TAATA | 618165 | 1.0137768 | 251.00153 | 2 |
| TTGTG | 379405 | 1.0107136 | 13.316986 | 3 |
| CGCGG | 170645 | 1.0100887 | 133.03853 | 45-49 |
| GAGTC | 286875 | 1.0099484 | 6.1438503 | 3 |
| TATAG | 479520 | 1.0040463 | 63.949875 | 15-19 |
| CCCCG | 160335 | 1.0025704 | 30.338316 | 1500-1591 |
| GGTAC | 281320 | 0.99039185 | 11.104713 | 1100-1199 |
| GGTAA | 368370 | 0.98826027 | 13.319865 | 1300-1399 |
| GTTGT | 363010 | 0.9670383 | 13.262675 | 2 |
| GGGAT | 280530 | 0.96089405 | 6.768372 | 50-59 |
| GTACC | 263385 | 0.9530328 | 9.300908 | 1400-1499 |
| CAACC | 255225 | 0.95253575 | 11.999573 | 1400-1499 |
| TAGGA | 354525 | 0.9511171 | 5.1850853 | 3 |
| AGTTG | 354100 | 0.94663393 | 13.636692 | 1 |
| TATCA | 431390 | 0.9283834 | 53.06715 | 30-34 |
| GTATC | 335275 | 0.921229 | 67.11313 | 30-34 |
| CAACG | 253175 | 0.91932404 | 82.54018 | 35-39 |
| GACAC | 251690 | 0.9139318 | 10.201467 | 6 |
| CCCGT | 191315 | 0.90841687 | 16.702826 | 1300-1399 |
| AACAT | 420615 | 0.9083916 | 5.7591934 | 9 |
| GTGTT | 340790 | 0.90784544 | 5.0835433 | 1 |
| GTTAC | 328775 | 0.90336895 | 13.05597 | 1200-1299 |
| GTGAC | 255935 | 0.90102357 | 7.88078 | 3 |
| GTTTA | 431750 | 0.90084136 | 13.788607 | 1500-1591 |
| AAACG | 324515 | 0.89797276 | 15.295963 | 1300-1399 |
| CGTAC | 248110 | 0.8977616 | 9.279555 | 1000-1099 |
| GTATT | 430265 | 0.89774305 | 9.988028 | 7 |
| ACGCA | 245120 | 0.8900749 | 89.84569 | 35-39 |
| TTGCA | 320475 | 0.8805632 | 10.99071 | 5 |
| AACGC | 240780 | 0.87431556 | 88.42427 | 35-39 |
| GTAAG | 322510 | 0.8652275 | 9.593502 | 2 |
| TACGA | 313060 | 0.86322695 | 420.4824 | 5 |
| TACTC | 304855 | 0.8609343 | 6.8170114 | 7 |
| CCCGG | 141225 | 0.8591872 | 36.935043 | 1500-1591 |
| TACGT | 312295 | 0.8580872 | 9.499411 | 1000-1099 |
| TGTAT | 405115 | 0.8452677 | 10.15818 | 6 |
| TAAGG | 315025 | 0.8451468 | 9.648197 | 3 |
| GGATT | 315045 | 0.8422261 | 10.355476 | 2 |
| GGTTA | 313380 | 0.83777493 | 17.571936 | 1300-1399 |
| ACGCG | 172705 | 0.80068505 | 102.60644 | 45-49 |
| ACGGG | 177330 | 0.79988724 | 24.934175 | 1300-1399 |
| CGGGA | 176985 | 0.798331 | 33.37636 | 45-49 |
| GGACG | 176555 | 0.7963914 | 37.49525 | 1500-1591 |
| ACGTA | 287545 | 0.7928722 | 7.9391003 | 1000-1099 |
| GTCGT | 226010 | 0.79287213 | 5.490606 | 900-999 |
| GTAAC | 287490 | 0.79272074 | 7.8704863 | 1300-1399 |
| GTTAA | 368505 | 0.7715968 | 14.418906 | 1500-1591 |
| GATCA | 276820 | 0.76329947 | 5.1156316 | 5 |
| TAACC | 268210 | 0.7601209 | 11.067327 | 1500-1591 |
| GTGTA | 284240 | 0.7598735 | 12.982497 | 5 |
| CCGGG | 128150 | 0.7585506 | 40.627026 | 1500-1591 |
| ATACG | 272985 | 0.7527247 | 420.76346 | 4 |
| GCAAC | 206390 | 0.7494393 | 9.742475 | 7 |
| TTAAC | 343745 | 0.73976487 | 15.883316 | 1500-1591 |
| ATACT | 342605 | 0.73731154 | 5.2387447 | 6 |
| TACGC | 202260 | 0.7318579 | 73.570206 | 45-49 |
| GGATC | 206870 | 0.72828937 | 5.899819 | 4 |
| AACGT | 259600 | 0.71581715 | 13.883002 | 1500-1591 |
| ACCCG | 148245 | 0.7063943 | 19.69334 | 1400-1499 |
| CGGGT | 157065 | 0.7059842 | 27.052168 | 1500-1591 |
| AACGG | 197860 | 0.6990295 | 22.582935 | 1300-1399 |
| GACGG | 153550 | 0.6926221 | 5.424509 | 1000-1099 |
| ACGGT | 194870 | 0.68604314 | 14.792024 | 1100-1199 |
| GATTG | 255830 | 0.68392366 | 10.300976 | 3 |
| ATTGC | 248880 | 0.683843 | 13.018717 | 9 |
| TATTG | 320735 | 0.66920996 | 9.971012 | 8 |
| CGACG | 143280 | 0.6642666 | 5.8163586 | 1000-1099 |
| ACCGT | 183315 | 0.66330725 | 12.677095 | 1200-1299 |
| CCGTT | 183625 | 0.6620908 | 17.627708 | 1400-1499 |
| ACGTT | 238105 | 0.6542367 | 12.756255 | 1500-1591 |
| ACGCC | 136545 | 0.6506433 | 22.20771 | 5 |
| CGTCG | 139800 | 0.64585197 | 5.011524 | 900-999 |
| CGGGC | 107700 | 0.6375021 | 16.80985 | 45-49 |
| CCGTA | 172510 | 0.6242105 | 8.8007145 | 1300-1399 |
| TACGG | 174620 | 0.61475277 | 15.25401 | 1100-1199 |
| CGTTT | 218505 | 0.5982693 | 15.228399 | 1500-1591 |
| GCCCG | 97250 | 0.5916512 | 101.143005 | 1500-1591 |
| GATAA | 270160 | 0.56767416 | 5.397768 | 1 |
| TTACG | 200530 | 0.5509925 | 11.157553 | 1100-1199 |
| CCGGT | 115305 | 0.5326893 | 18.627483 | 1400-1499 |
| CGTAA | 191330 | 0.5275704 | 7.3239217 | 1100-1199 |
| TACCG | 143995 | 0.52103174 | 12.412991 | 1200-1299 |
| CTACG | 143530 | 0.5193492 | 5.1814747 | 1500-1591 |
| CGGTA | 146295 | 0.5150341 | 10.740994 | 1200-1299 |
| CGGTT | 146750 | 0.51481783 | 18.859562 | 1500-1591 |
| CTCGG | 110210 | 0.5091513 | 7.534968 | 1 |
| AACCG | 138685 | 0.5035902 | 21.844633 | 1500-1591 |
| CCGGA | 105845 | 0.49071258 | 5.9464192 | 1500-1591 |
| CGCCA | 102855 | 0.49010882 | 21.0422 | 6 |
| ACCGG | 104785 | 0.48579824 | 21.029924 | 1400-1499 |
| CGTTA | 173780 | 0.4774921 | 12.374091 | 1200-1299 |
| GGCGC | 71060 | 0.4206212 | 5.840862 | 7 |
| TAACG | 149055 | 0.411002 | 13.325021 | 1500-1591 |
| GACGT | 115965 | 0.4082568 | 29.26417 | 1500-1591 |
| TCGGA | 84330 | 0.29688522 | 5.756271 | 2 |
| CGGAT | 66300 | 0.23341027 | 13.924314 | 1 |