![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 6494_GGCTAC_R1.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 131737035 |
| Filtered Sequences | 0 |
| Sequence length | 100 |
| %GC | 47 |
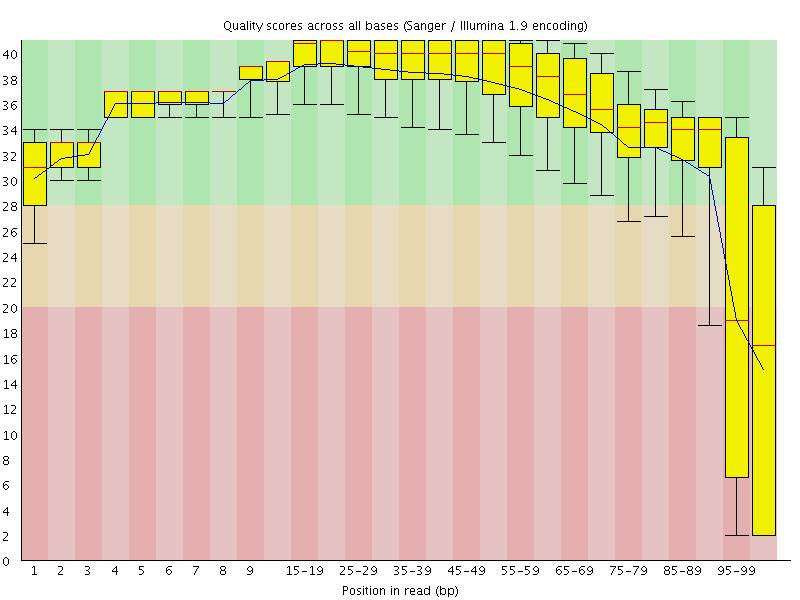
![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality
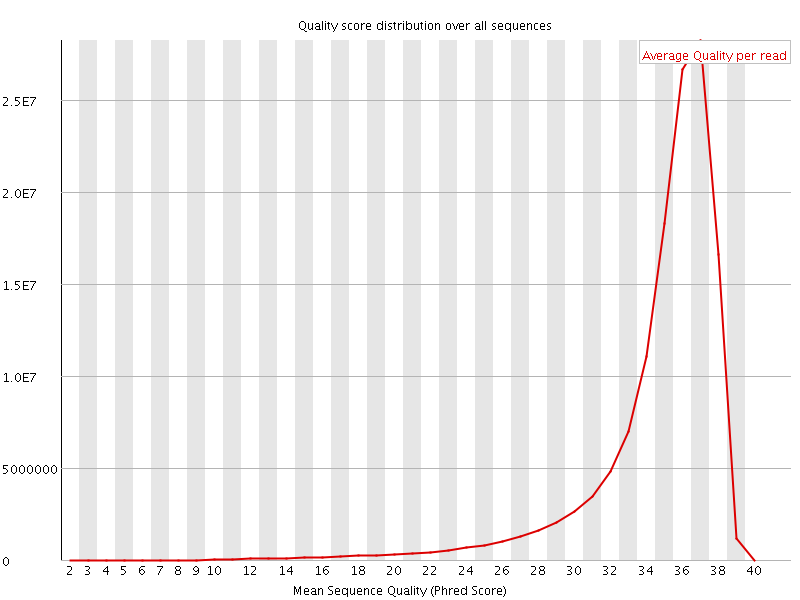
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
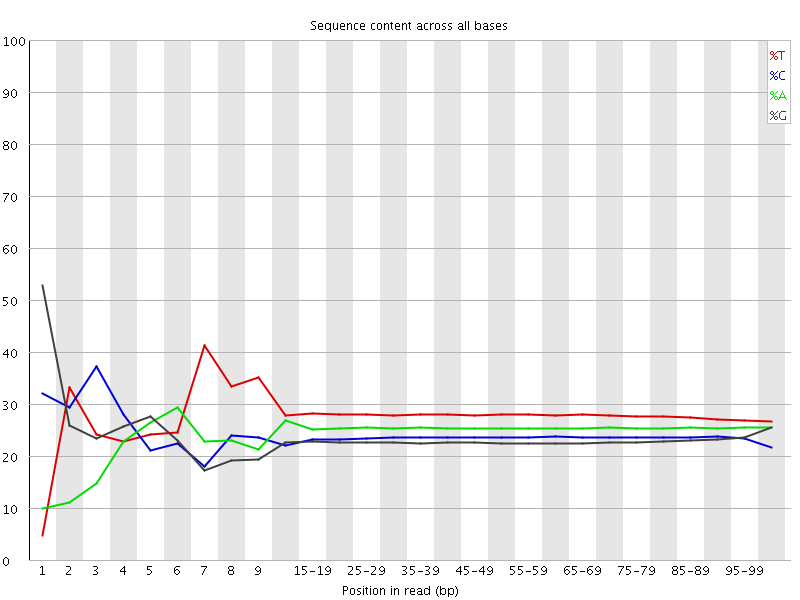
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
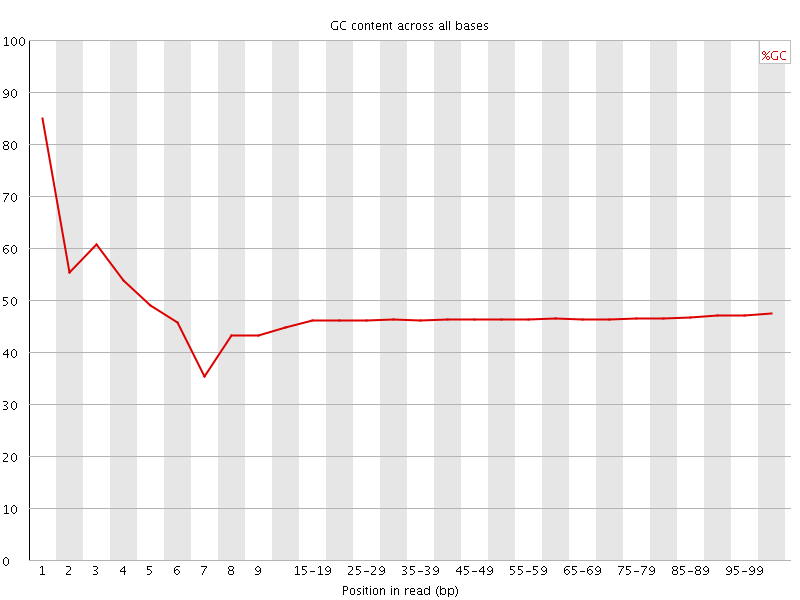
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
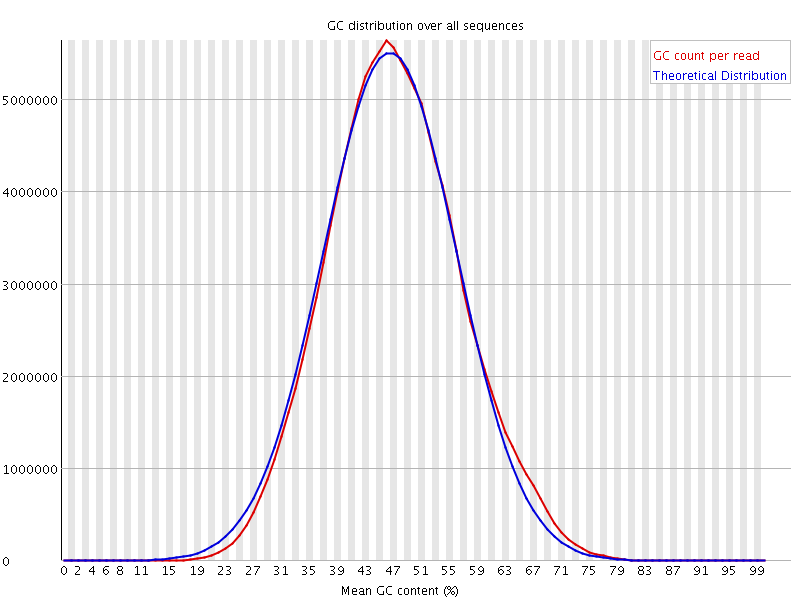
![[WARN]](Icons/warning.png) Per base N content
Per base N content
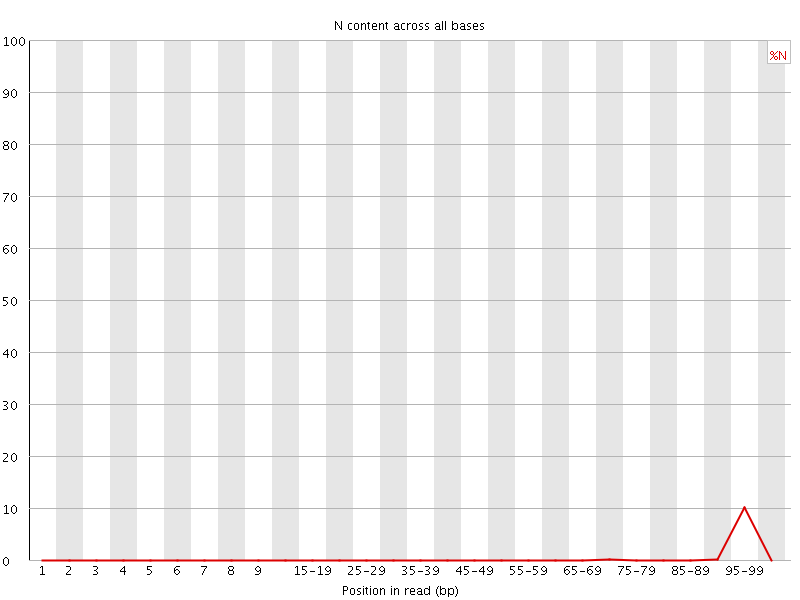
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
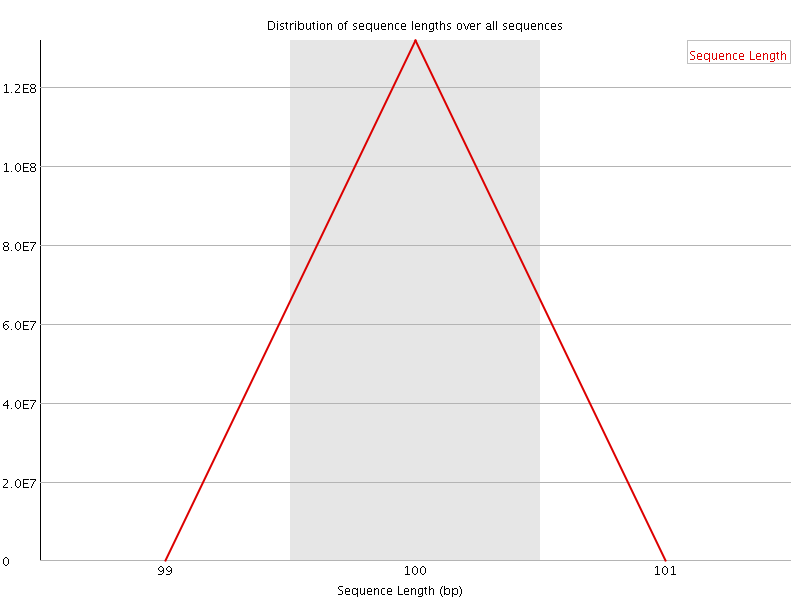
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
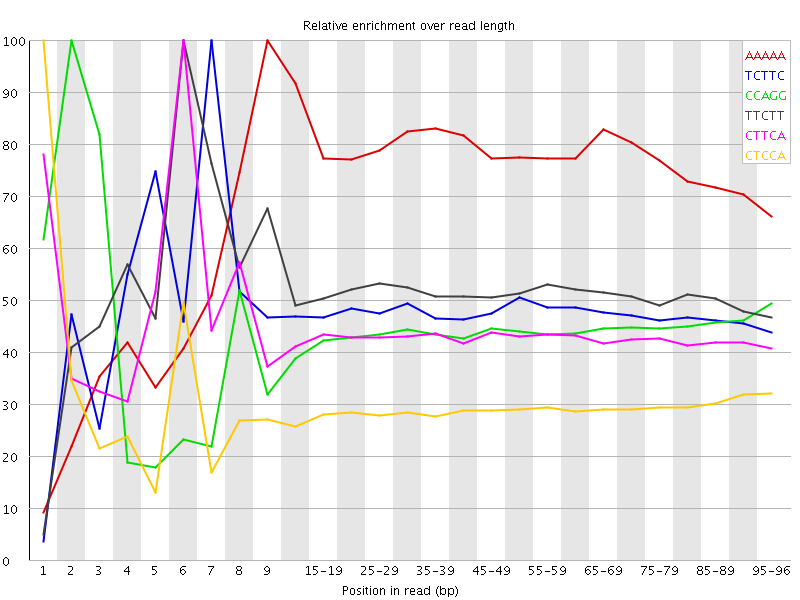
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 41084735 | 3.3300562 | 4.409197 | 9 |
| TCTTC | 42831025 | 2.8338983 | 5.9311852 | 7 |
| CCAGG | 24556130 | 2.602913 | 5.9029937 | 2 |
| TTCTT | 45564045 | 2.5800562 | 5.011365 | 6 |
| CTTCA | 33930060 | 2.48821 | 5.7084527 | 6 |
| CTCCA | 28788355 | 2.4668236 | 8.364095 | 1 |
| CCAGC | 23195945 | 2.3896372 | 5.8898244 | 2 |
| CAGCT | 27099850 | 2.389285 | 5.535928 | 3 |
| TCCAG | 26764945 | 2.359758 | 6.7397556 | 2 |
| GGCAG | 21186040 | 2.3106241 | 11.347821 | 1 |
| CAGGA | 22940785 | 2.3065653 | 5.1073995 | 3 |
| CTCCT | 29393495 | 2.2724578 | 5.8940825 | 1 |
| CTGCT | 27457950 | 2.1842012 | 5.4597697 | 3 |
| GCAGG | 19730160 | 2.1518407 | 5.097575 | 2 |
| GCCAG | 20182250 | 2.1392882 | 11.78663 | 1 |
| GGCTG | 21574035 | 2.1229231 | 8.415544 | 1 |
| GCTGG | 21365105 | 2.102364 | 6.440885 | 1 |
| CCCAG | 20121255 | 2.0728838 | 6.9681864 | 1 |
| CCACA | 21781150 | 2.06861 | 5.5726776 | 2 |
| CCTGG | 20152840 | 1.9273447 | 5.1297736 | 1 |
| GGCCA | 17718620 | 1.8781471 | 8.062006 | 1 |
| GGGCA | 16828870 | 1.8354161 | 7.0198064 | 1 |
| GGGGG | 14962795 | 1.8213533 | 9.535551 | 1 |
| CTCTG | 22885265 | 1.8204567 | 5.1649933 | 1 |
| CCAGA | 18571710 | 1.8148032 | 5.4994316 | 2 |
| GGGAG | 16107430 | 1.8075305 | 6.7470956 | 1 |
| GGGGC | 14181965 | 1.6777917 | 6.882024 | 1 |
| GCTCC | 17765260 | 1.6512576 | 5.455988 | 1 |
| GGGGA | 14666750 | 1.6458615 | 7.157799 | 1 |
| GCCAC | 15708705 | 1.6183046 | 8.099069 | 1 |
| CCCCA | 15742710 | 1.5762299 | 5.640662 | 1 |
| GCCTG | 15910400 | 1.5216131 | 7.134165 | 1 |
| GGCCT | 15606395 | 1.4925389 | 5.4717517 | 1 |
| GCCTC | 15972420 | 1.4846156 | 6.209764 | 1 |
| GTGGG | 14455640 | 1.4635923 | 6.394421 | 1 |
| GGGGT | 13968700 | 1.4142909 | 7.0125785 | 1 |
| GTCCA | 16003885 | 1.4109983 | 9.290385 | 1 |
| GCCCA | 13569275 | 1.3979014 | 7.1524243 | 1 |
| GCCAA | 14127335 | 1.3805047 | 5.572314 | 1 |
| GGCAT | 14935040 | 1.3548375 | 5.54831 | 1 |
| GTCTG | 16460980 | 1.3472865 | 6.1431975 | 1 |
| GCCTT | 16855925 | 1.3408402 | 5.2929497 | 1 |
| GGCAA | 13097980 | 1.3169273 | 5.3036137 | 1 |
| GCCAT | 14838665 | 1.3082654 | 5.7831726 | 1 |
| GTCAG | 14006695 | 1.2706226 | 6.8097825 | 1 |
| GTCCT | 15289750 | 1.2162557 | 7.1161427 | 1 |
| GCCCC | 10822905 | 1.175455 | 5.378905 | 1 |
| GCCCT | 11604615 | 1.0786338 | 5.526143 | 1 |
| GTCCC | 10414100 | 0.967977 | 5.217869 | 1 |