![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 6491_ATCACG_R2.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 135218638 |
| Filtered Sequences | 0 |
| Sequence length | 100 |
| %GC | 48 |
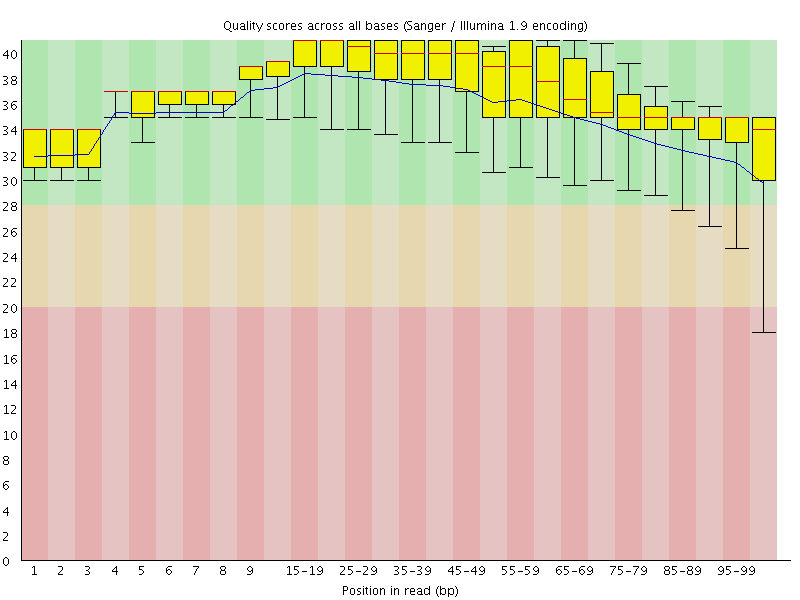
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
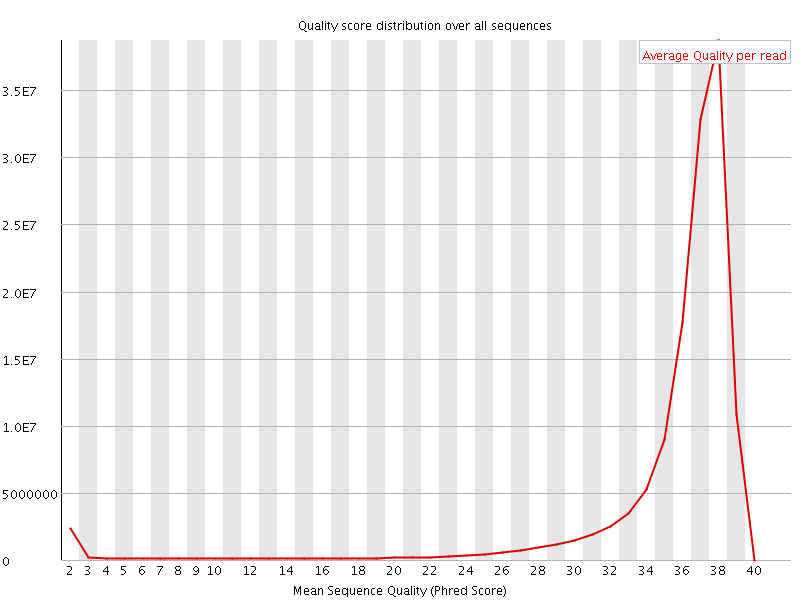
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
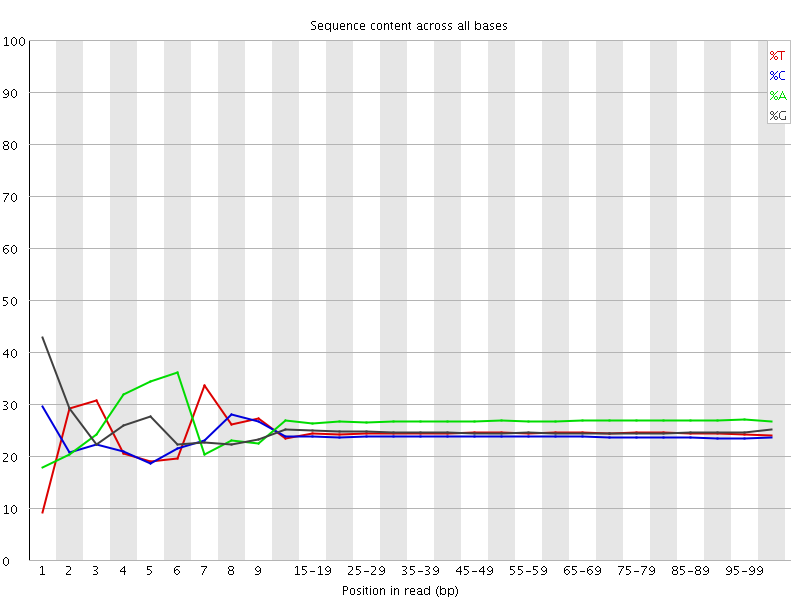
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
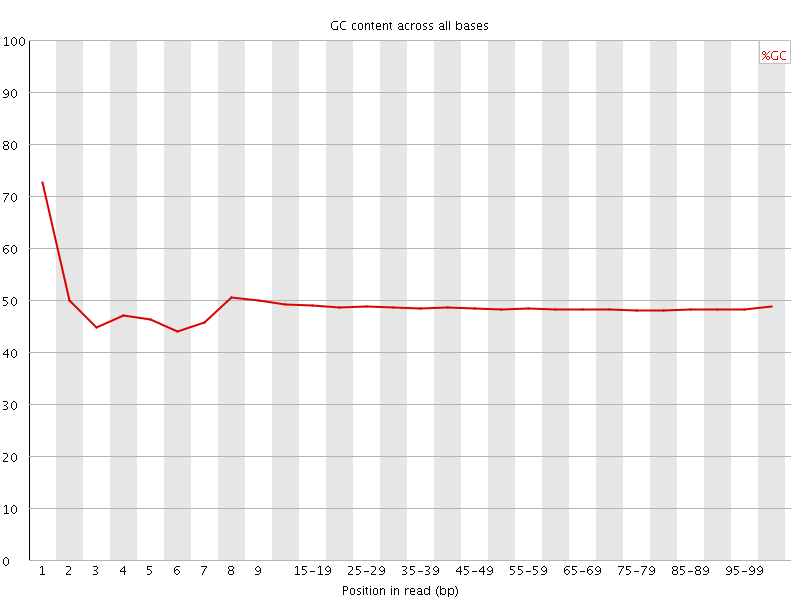
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
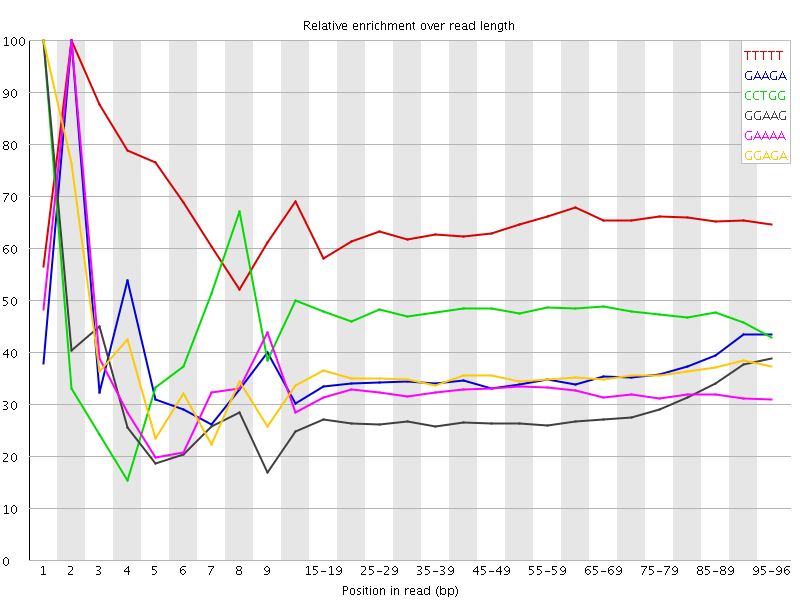
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 49503135 | 4.3371134 | 6.6650314 | 2 |
| GAAGA | 44191025 | 2.8659708 | 7.9414134 | 2 |
| CCTGG | 31024650 | 2.7754025 | 5.846671 | 1 |
| GGAAG | 30707140 | 2.1439562 | 7.3905888 | 1 |
| GAAAA | 34017305 | 2.0492682 | 6.2348623 | 2 |
| GGAGA | 28925955 | 2.019595 | 5.558608 | 1 |
| CTTCA | 22665640 | 1.9135994 | 6.126926 | 1 |
| GGAAA | 28164715 | 1.8265985 | 6.8593984 | 1 |
| GTGGA | 23683215 | 1.8086679 | 5.017961 | 1 |
| CCTGA | 21450790 | 1.7824754 | 5.024982 | 1 |
| AAGAT | 24927735 | 1.642568 | 5.0888247 | 3 |
| GTTTT | 18219470 | 1.5710899 | 5.721496 | 1 |
| GTTTG | 14945555 | 1.2684535 | 5.688326 | 1 |